En atención a la creciente preocupación sobre la confianza en...
Leer más
SARS-CoV-2 viral load and shedding kinetics
Abstract
SARS-CoV-2 viral load and detection of infectious virus in the respiratory tract are the two key parameters for estimating infectiousness. As shedding of infectious virus is required for onward transmission, understanding shedding characteristics is relevant for public health interventions. Viral shedding is influenced by biological characteristics of the virus, host factors and pre-existing immunity (previous infection or vaccination) of the infected individual. Although the process of human-to-human transmission is multifactorial, viral load substantially contributed to human-to-human transmission, with higher viral load posing a greater risk for onward transmission. Emerging SARS-CoV-2 variants of concern have further complicated the picture of virus shedding. As underlying immunity in the population through previous infection, vaccination or a combination of both has rapidly increased on a global scale after almost 3 years of the pandemic, viral shedding patterns have become more distinct from those of ancestral SARS-CoV-2. Understanding the factors and mechanisms that influence infectious virus shedding and the period during which individuals infected with SARS-CoV-2 are contagious is crucial to guide public health measures and limit transmission. Furthermore, diagnostic tools to demonstrate the presence of infectious virus from routine diagnostic specimens are needed.
Introduction
At the end of 2019, a novel coronavirus emerged, later termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19). SARS-CoV-2 primarily targets multiciliated cells in the upper respiratory tract (URT), but was also reported to infect cells outside the URT1. It can spread to the lower respiratory tract (LRT), where it infects alveoli, leading to reduced gas exchange, inflammation and pulmonary pathologies that are typical of COVID-19 (ref.2). Individuals who are infected shed the virus through the URT, with emission of infectious virus leading to secondary transmission and thus further spread of the virus.
Because of their nonspecific clinical presentation, precise diagnostic tools are needed to identify SARS-CoV-2 infections. Specific real-time PCR (RT-PCR) assays were quickly available after the emergence of the virus, later followed by antigen-detecting (rapid) diagnostic tests (Ag-RDTs) and serological assays. Although detection of viral RNA in respiratory specimens by RT-PCR is highly sensitive and specific, it does not distinguish between replication-competent virus and residual RNA. In the absence of a diagnostic test, infectiousness is often established using one of two proxies: the presence of viral RNA above a defined cycle threshold (Ct) value, or a positive Ag-RDT. RT-PCR is a useful tool for initial diagnosis, whereas Ag-RDTs can serve as an indicator for ending the isolation period. This is because viral RNA (which would be picked up by RT-PCR) remains detectable in the absence of infectious virus, whereas positivity of Ag-RDTs better correlates with the presence of infectious virus.
Aside from the respiratory tract, SARS-CoV-2 RNA has been detected in peripheral blood, stool, urine and ocular secretions3,4,5,6,7. Virus isolation from non-respiratory specimens was unsuccessful in most studies4,8,9, with very few reported cases of infectious virus presence in non-respiratory specimens10,11,12,13. Furthermore, viral loads from respiratory tract samples were found to be much higher than from other materials, the latter often with RNA viral loads that are incompatible with the presence of infectious virus. Such specimens are not considered relevant for transmission and therefore, we concentrate on SARS-CoV-2 virus shedding only through the respiratory tract.
Here, we elucidate the relationship between SARS-CoV-2 viral load and infectious virus presence, the biological and host factors that determine infectious virus shedding, measurement of infectious virus and the role diagnostics can have as a proxy for infectious virus shedding.
Measuring SARS-CoV-2 viral load
The gold standard for laboratory diagnosis of a respiratory tract infection is demonstration of viral RNA with a virus-specific (semi-)quantitative RT-PCR from material collected from the respiratory tract. The most commonly used materials are swab specimens from the nasopharynx or oropharynx, but swabs of the nasal cavity, saliva or gargled liquid solution have also been suggested as alternative materials, with the advantage of being a less uncomfortable procedure for the participant. Viral load as determined by RT-PCR is either expressed as the number of viral RNA copies per millilitre of viral transport medium or per swab, or by the arbitrary test-specific Ct value. By contrast, infectiousness is determined by qualitative or quantitative assessment of infectious virus in a clinical specimen by replication of virus in cell culture. The limitations to measuring viral shedding are described in Box 1. In this Review, we refer to viral particles that can cause infection as infectious virus, and to viral RNA levels (which are widely used as surrogates for infectious virus) as viral load.
Detection of infectious virus
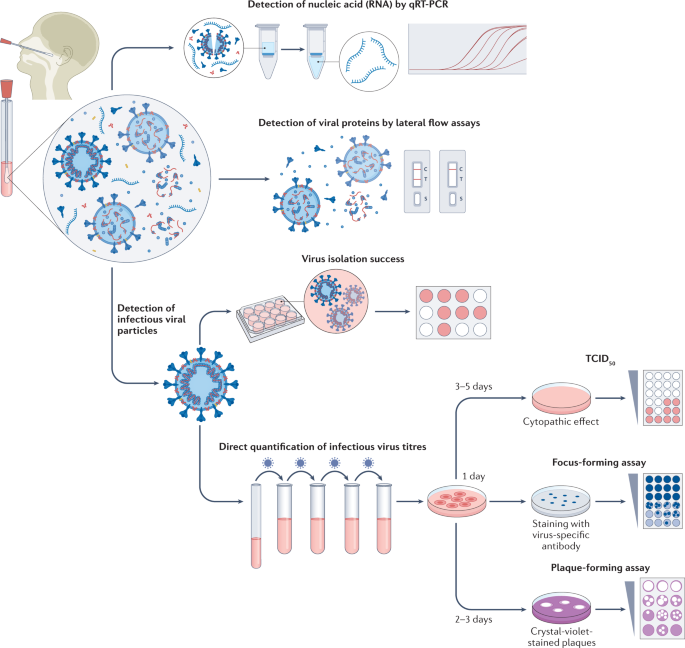
The gold standard for determining the presence of infectious (that is, replication competent) virus in respiratory specimens is the recovery of virus in cell culture, a procedure that is commonly termed virus isolation (Fig. 1).
Swab specimens from the nasopharynx or oropharynx are used for detection of SARS-CoV-2 viral loads. Detection of viral nucleic acids (RNA) is performed by quantitative real-time PCR (qRT-PCR). Viral RNA is extracted from lysed virus, reverse transcribed and amplified by qPCR using primers specific for one or more target regions in the viral genome. The amplification cycle at which samples cross the threshold (cycle threshold) defines the amount of viral RNA. RNA viral load can be expressed as the number of viral RNA copies per millilitre, or by the arbitrary test-specific cycle threshold value. Lateral flow assays detect the presence of specific viral proteins in the lysed viral particles. SARS-CoV-2 nucleocapsid is used in most antigen-detecting (rapid) diagnostic tests. The presence of infectious (replication-competent) virus in respiratory specimens can only be determined by the recovery of virus in cell culture by isolation or by quantification of infectious virus titres using 50% tissue culture infectious dose (TCID50), focus-forming assays or plaque-forming assays. Virus isolation is performed by applying infectious medium on the monolayer of cells; isolation success is determined by the presence of a cytopathic effect approximately 3–5 days post-infection. White colour indicates the presence of a cytopathic effect in cells. For quantification of infectious virus titres, serial dilutions of respiratory samples are performed and used for inoculation on the monolayer of cells. In TCID50, 3–5 days post-infection, viral-induced cytopathic effect is classically defined using microscopy. In focus-forming assays, cells are fixed 1 day post-infection and immunostaining with virus-specific antibodies is performed to detect groups of infected cells (foci). The foci, indicating the presence of infectious virus, are displayed in blue. In plaque-forming assays, plates are fixed 2–3 days post-infection and stained with crystal violet; wells with individual plaques are used to determine viral titres. The plaques, indicating the presence of infectious virus, are displayed in white.
In the case of SARS-CoV-2, various cell lines and primary cells can be used for virus isolation, including those that express angiotensin-converting enzyme 2 (ACE2; the receptor required for virus entry) or transmembrane protease 2 (TMPRSS2; which is also important for virus entry)14. A cell line derived from African green monkey kidney cells, Vero E6, is commonly used for virus isolation, propagation and titration15. Other human cell lines that have been successfully used for SARS-CoV-2 isolation are a colorectal adenocarcinoma cell line (Caco-2), a lung adenocarcinoma cell line (Calu-3), a lung adenocarcinoma cell line ectopically overexpressing ACE2 (A549) and a human hepatocellular carcinoma cell line (Huh7)16,17.
The presence of infectious virus in the cell culture is qualitatively assessed using light microscopy, which can be used to identify cells undergoing the cytopathic effects (and death) caused by SARS-CoV-2 infection, consisting of syncytium formation, cell rounding, detachment and degeneration17. Infection is usually confirmed by a second method, either by a specific RT-PCR for viral RNA from the supernatant of infected cells, indicating virus replication by an increase of viral load over time in comparison to the baseline sample, or by immunostaining for viral proteins15,18.
This qualitative measurement of virus presence cannot, however, quantify the infectious virions in the inoculated specimens, although samples with lower viral load commonly show delayed development of a cytopathic effect19. Instead, methods such as plaque assays, focus-forming assays or 50% tissue culture infectious dose (TCID50) can be used to quantify infectious virus in a patient sample.
The above methodologies are reliable tools to detect infectious virus in clinical specimens of individuals who are infected with SARS-CoV-2, although there are limitations. Detection of viable virus particles is highly influenced by the quality of the sample, and infectious viral particles can quickly lose their infectiousness in unsuitable storage conditions. To preserve infectious virus in specimens, swab samples from patients infected with SARS-CoV-2 should be immediately submerged in a viral transport medium suitable for cell culture and stored at −80 °C as early as possible after collection. Prolonged exposure to higher temperatures or repeated freeze–thaw cycles can drastically influence the quality of the sample, leading to potentially complete loss of infectious viral particles. Therefore, many factors can influence the reproducibility of the results between different laboratories. Furthermore, cell lines used for isolation can show a high variability between laboratories even when they are presumably the same. Consumables used during cell culture, such as culture medium or additives such as fetal bovine serum and antibiotics, could potentially also impact virus isolation success. In human primary airway epithelial cells, which mimic the primary site of entry in the human respiratory tract, the probability of isolating infectious virus was reduced compared with that of Vero E6 cells, indicating that infectious virus determined using Vero E6 cells might be overestimated for assessing transmission risks in vivo20.
Importantly, all cell culture work with SARS-CoV-2 is done under biosafety level 3 conditions, so only specially trained personnel in laboratories with advanced infrastructure can perform these experiments. Thus, detection of viable virus through virus isolation is not suitable for diagnostics and is restricted to research only.