En atención a la creciente preocupación sobre la confianza en...
Leer más
Structures of the Omicron Spike trimer with ACE2 and an anti-Omicron antibody
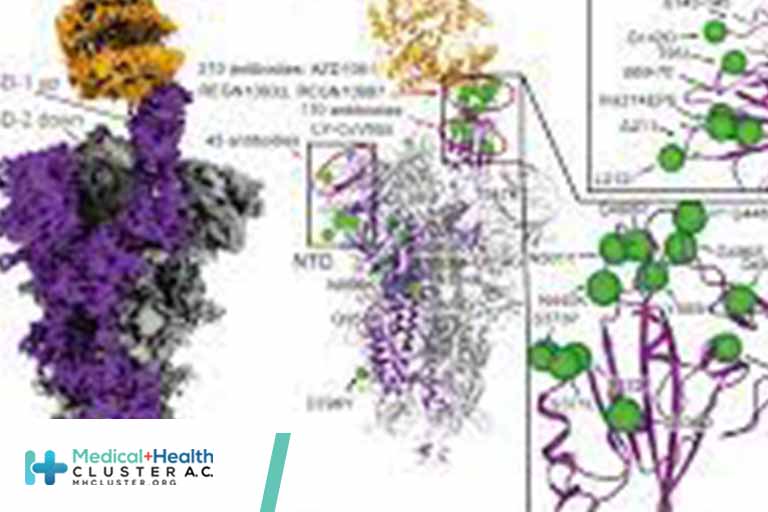
The SARS-CoV-2 Omicron variant has become the dominant infective strain. We report the structures of the Omicron spike trimer on its own or in complex with ACE2 or an anti-Omicron antibody. Most Omicron mutations are located on the surface of the spike protein, which change binding epitopes to many current antibodies. In the ACE2 binding site, compensating mutations strengthen RBD binding to ACE2. Both the RBD and the apo form of the Omicron spike trimer are thermodynamically unstable. An unusual RBD-RBD interaction in the ACE2-spike complex supports the open conformation and further reinforces ACE2 binding to the spike trimer. A broad-spectrum therapeutic antibody, JMB2002, which has completed a Phase 1 clinical trial, maintains neutralizing activity against Omicron. JMB2002 binds to RBD differently from other characterized antibodies and inhibits ACE2 binding.
The Omicron variant of SARS-CoV-2, the causative virus of COVID-19, was initially reported from South Africa in November 2021, and quickly became the dominant strain worldwide (1). Phylogenetic tree analyses reveal that Omicron evolved independently from previous variants of concerns (VOC), including the predominant Alpha, Beta, Gamma, and Delta variants (Fig. 1A) (2–5). Compared to the original wildtype (WT) strain of SARS-CoV-2, Omicron has 60 amino acid mutations, of which 37 mutations are in the spike protein, the target of most COVID-19 vaccines and therapeutic antibodies (Fig. 1B). This high variation is reflected in different behavior with the Omicron variant showing enhanced transmission, antibody evasion, and vaccine resistance (6–8).

To study the mechanism for Omicron’s enhanced transmission, we first biochemically characterized the interactions of the SARS-CoV-2 receptor ACE2 with the trimer of the spike extracellular domain (ECD) from Omicron and the original WT strain, both of which contain proline substitutions (2P or 6P) and a mutated furin cleavage site to stabilize the prefusion conformation (9, 10). Monomeric human ACE2 bound to immobilized Omicron trimeric spike protein with approximately 6-fold higher affinity (KD=2.5 ± 0.6 nM) than WT spike trimer (KD=14.7 ± 4.9 nM). The dimeric human ACE2 bound to immobilized biotinylated Omicron spike trimer (KD=0.3 ± 0.2 nM) with approximately 9-fold higher avidity than WT (KD=2.7 ± 1.4 nM) (Fig. 1, C and D). We then studied the interactions of ACE2 with monomeric receptor binding domain (RBD) from Omicron and WT strains. Monomeric human ACE2 bound to immobilized Omicron RBD (KD=38.9 ± 10.5 nM) with approximately 2-fold higher affinity than WT RBD (KD=75.5 ± 2.1 nM) (Fig. 1, C and D). The enhanced interaction of Omicron spike and RBD proteins with human ACE2 is consistent with previously published data (11), and may contribute to the increased infectivity of the Omicron variant.
To determine the structural basis of higher affinity of the Omicron spike trimer for ACE2, we solved the structure of the ACE2-Omicron spike trimer complex at a global resolution of 2.77 Å (table S1). Despite an excess of ACE2 (molar ratio of 3.2 ACE2 to 1 spike trimer; fig. S1A), we only observed strong density for one ACE2 bound to one RBD from the spike trimer in the open “up” conformation (Fig. 2A and fig. S2). The other two RBDs, with clear density, are in the closed “down” conformation. Particle classification revealed that the majority of picked particles (~70%) do not have ACE2 bound. We also determined the structure of this apo Omicron spike trimer at a global resolution of 2.56 Å (fig. S2 and table S1). All three RBDs are in the closed down conformation but they are less visible in the high-resolution map (2.56 Å; fig. S3A), yet become more visible in lower resolution maps (4.5 Å and 6.5 Å; fig. S3, B and C). This contrasts with the clear visibility of the three RBDs in the ACE2-Omicron spike complex in a high-resolution map (2.56 Å; Fig. 2A), indicating that the RBD in the apo form is more dynamic and ACE2 binding likely stabilizes the conformation of the three RBDs. Thermal shift assays at pH7.4 revealed that the Omicron and WT RBD have single melting temperatures of 45.7°C and 51.0°C respectively (fig. S1C), indicating that the Omicron RBD is less stable than the WT RBD. In contrast, both the Omicron and WT spike trimer displayed two melting temperatures (fig. S1D), with the high Tm corresponding to the dissociation of the spike trimer and the low Tm corresponding to unfolding of the RBD. The melting temperature profile of WT spike trimer is similar to previous reports (10, 12). Tm1 of both Omicron and WT spike trimer is similar to the respective Tm for the isolated RBD (fig. S1, C and D), indicating that the Omicron RBD within the context of the spike trimer remains less stable than the WT RBD. We further confirmed the highly flexible nature of the Omicron RBD by performing hydrogen-deuterium exchange mass spectrometry (HDX), which showed that the Omicron spike trimer has an overall higher rate of HDX (fig. S4), particularly in the RBD region, consistent with its lower thermal stability.
Créditos: Comité científico Covid