En atención a la creciente preocupación sobre la confianza en...
Leer más
SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex
Abstract
The newly reported Omicron variant is poised to replace Delta as the most prevalent SARS-CoV-2 variant across the world. Cryo-EM structural analysis of the Omicron variant spike protein in complex with human ACE2 reveals new salt bridges and hydrogen bonds formed by mutated residues R493, S496 and R498 in the RBD with ACE2. These interactions appear to compensate for other Omicron mutations such as K417N known to reduce ACE2 binding affinity, resulting in similar biochemical ACE2 binding affinities for Delta and Omicron variants. Neutralization assays show that pseudoviruses displaying the Omicron spike protein exhibit increased antibody evasion. The increase in antibody evasion, together with retention of strong interactions at the ACE2 interface, thus represent important molecular features that likely contribute to the rapid spread of the Omicron variant.
The Omicron (B.1.1.529) variant of SARS-CoV-2, first reported in November 2021, was quickly identified as a variant of concern with potential to spread rapidly across the world. This concern is heightened as the Omicron variant is currently circulating even amongst doubly vaccinated individuals. SARS-CoV-2 relies on a trimeric spike protein for host cell entry via recognition of the angiotensin converting enzyme 2 (ACE2) receptor. The Omicron variant spike protein has 37 mutations as compared to 12 mutations in the Gamma variant, the most mutated variant to emerge before Omicron (1). Understanding the consequences of these mutations for ACE2 receptor binding and neutralizing antibody evasion is important in guiding the development of effective therapeutics to limit the spread of the Omicron and related variants.
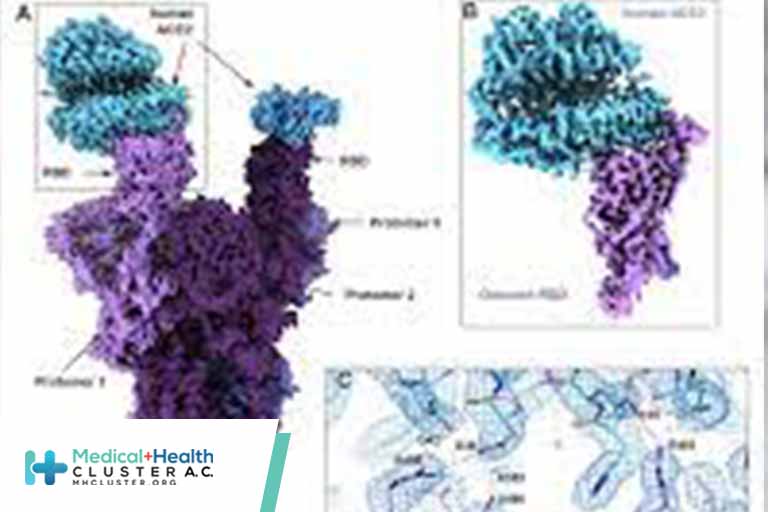
The spike protein comprises 2 domains, the S1 domain, which contains the receptor binding domain (RBD) and the S2 domain responsible for membrane fusion. The Omicron variant has 37 mutations (Fig. 1A) in the spike protein relative to the initial Wuhan-Hu-1 strain, with 15 of them present in the receptor binding domain (RBD) (1). The RBD mediates attachment to human cells through the ACE2 receptor and is the primary target of neutralizing antibodies (2, 3). The Delta variant, which was the predominant SARS-CoV-2 lineage until the emergence of Omicron, has 7 mutations in the spike protein relative to the Wuhan-Hu-1 strain, with 2 mutations falling within its RBD. Of the Delta spike mutations, two (T478K in the RBD and D614G at the C terminus of S1) are shared with the Omicron strain. Analysis of the sequence of the Omicron genome suggests that it is not derived from any of the currently circulating variants, and may have a different origin (4).
Cryo-EM structural analysis of the Omicron spike protein ectodomain shows that the overall organization of the trimer is similar to that observed for the ancestral strain (5–7) and all earlier variants (8–10) (Fig. 1B and table S1). The RBD in one of the protomers (protomer 1) is well-resolved and is in the “down” position, while the other two RBDs are less well-resolved because they are flexible relative to the rest of the spike protein polypeptide. Similarly, the amino terminal domain (NTD) is poorly resolved, reflecting the dynamic and flexible nature of this domain. The mutations in the Omicron variant spike protein are distributed both on the surface and the interior of the spike protein (Fig. 1C), including NTD and RBD regions. The mutations in the RBD are predominantly distributed on one face of the domain (Fig. 1D), which spans regions that bind ACE2 as well as those that form epitopes for numerous neutralizing antibodies (11).
The Omicron variant shares RBD mutations in common with previous variants of concern (K417N, T478K, and N501Y). The N501Y and K417N mutations impart increased and decreased ACE2 binding affinities respectively (8, 12–16). These mutational effects preserve the same general impact on ACE2 affinity when present in isolation or in combination with other RBD mutations (12). However, the Omicron RBD contains additional mutations, the majority of which have been shown to decrease receptor binding in a high-throughput assay (table S2) (17), with the exception of G339D, N440K, S447N, and Q498R (17, 18). To measure the impact of Omicron spike protein mutations on human ACE2 binding affinity, we performed surface plasmon resonance studies and compared the resulting apparent binding affinities (KD,app) to wild-type and Delta spikes (Fig. 2). Wild-type (WT) is used in this work to refer to the ancestral Wuhan-Hu-1 strain with the addition of the D614G mutation. While the Omicron spike protein exhibits a measurable increase in apparent affinity for ACE2 relative to the wild-type spike [in agreement with a recent preprint (19)], the apparent ACE2 affinity is similar for Delta and Omicron variants (Fig. 2D). Despite harboring several RBD mutations which decrease ACE2 binding (fig. S2) (12, 16, 17), the preservation of overall ACE2 binding affinity for the Omicron spike protein suggests there are compensatory mutations that restore higher affinity for ACE2. Such mutational effects should be possible to visualize in a high-resolution structure of the spike protein-ACE2 complex.
Créditos: Comité científico Covid